Antiparasitic peptides discovery
January 14, 2023
I contributed to this project as part of my work as research assistant at the Applied Signal Processing and Machine Learning Group and the and the Grupo de Medicina Molecular y Traslacional - Universidad San Francisco de Quito, in Ecuador.
Summary
Antimicrobial peptides (AMPs) are small bioactive chemicals that have appeared as promising compounds to treat a wide range of diseases. The effectiveness of AMPs resides in the wide range of mechanisms they can use for both killing microbes and modulating immune responses. However, the AMPs’ chemical space (AMPCS) is huge, it is estimated that there exist more than 10^65 unique sequences of peptides with 50 residues or fewer, which represent a big challenge for the discovery of new promising sequences and the identification of common features, motifs, or relevant biological functions shared by these peptides. Therefore, we present a new approach based on network science and similarity searching to discover new potential AMPs, specifically antiparasitic peptides (APPs). The graphical summary of this research is presented below.

Figure 1.- Graphical Abstract of the Antiparasitic peptides discovery project. Retrieved from Ayala-Ruano, et al., 2021, ChemRxiv.
We have taken advantage of network-based representation of APPs’ chemical space (APPCS) to retrieve valuable information, using three types of networks: chemical space (CSN), half-space proximal (HSPN), and metadata (METN).

Figure 2.- Networks of the Antiparasitic peptides discovery project. Modified from Ayala-Ruano, et al., 2021, ChemRxiv.
Some centrality measures were applied to identify the most important and non-redundant nodes, and these peptides were taken as queries against the graph database starPepDB to discover new potential APPs with similarity searching by group fusion (MAX-SIM rule) models. We evaluated the multi-query similarity searching models (mQSSMs) performance with five benchmarking data sets of APP/non-APPs. It can be stated that the predictions performed by the best mQSSMs presented a strong-to-very strong predictive agreement since their external Matthews correlation coefficient values ranged from 0.834 to 0.965. Then, we compared the performance metrics of our mQSSMs with machine learning APP prediction servers AMPDiscover and AMPFun.The model proposed in this report outperformed the machine learning approaches with statistically significant differences, showing the enormous potential of this method.
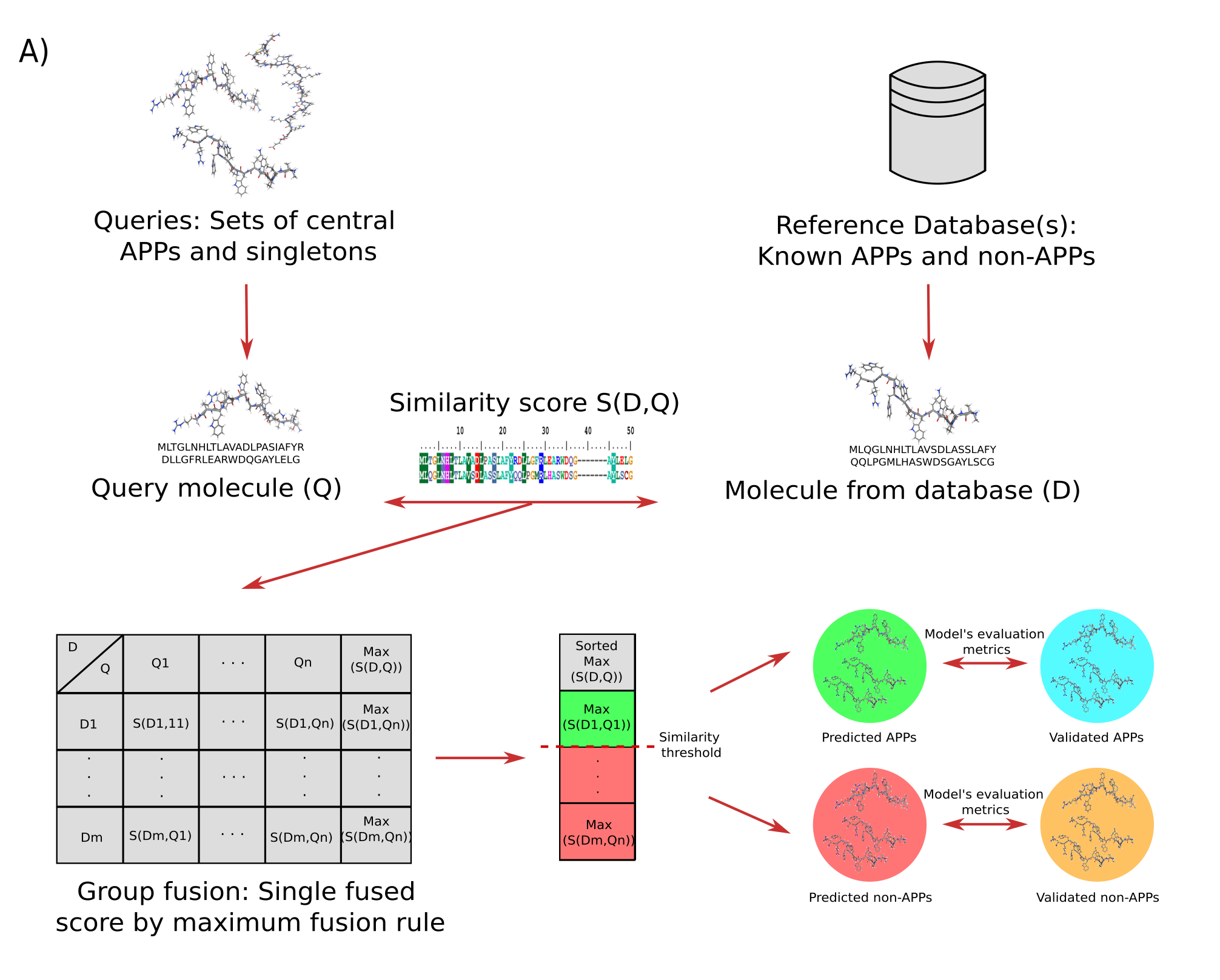
Figure 3.- Workflow corresponding to the similarity searching modeling process. Modified from Ayala-Ruano, et al., 2021, ChemRxiv.
After applying our method and additional filters, we proposed 95 repurposed leads as potential APPs, which have not been associated with this activity until now. In addition, we explored sequence similarities and motifs shared by these peptides, which can serve as templates for searching and designing new promising APPs. The analyses showed that the similarity models proposed in this study could contribute to identifying APPs with high effectivity and reliability. Our models and pipeline are freely available through the starPep toolbox software.

Figure 4.- Filtering workflow to obtain the new potential APPs. Obtained from Ayala-Ruano, et al., 2021, ChemRxiv.
Citation
Ayala-Ruano S., Marrero-Ponce Y., Aguilera‑Mendoza L., Pérez N., Agüero-Chapin G., Antunes A., Aguilar A. (2022). Exploring the Chemical Space of Antiparasitic Peptides and Discovery of New Promising Leads through a Novel Approach based on Network Science and Similarity Searching. ACS omega. doi: doi.org/10.1021/acsomega.2c03398.
- Posted on:
- January 14, 2023
- Length:
- 3 minute read, 514 words
- Tags:
- Antimicrobial peptides
- See Also:
- Prediction of antimicrobial peptides using machine learning classifiers
- Exploring the Chemical Space of Antiparasitic Peptides and Discovery of New Promising Leads through a Novel Approach based on Network Science and Similarity Searching
- Exploring the Chemical Space of Antiparasitic Peptides and Discovery of New Promising Leads through a Novel Approach based on Network Science and Similarity Searching