CRBN mutations
September 7, 2021
I worked on this project during the Global Intern Program 2019, a summer research internship at the Tumor Metabolism and Therapeutic Oncology Laboratory - Gwangju Institute of Science and Technology, in South Korea. Also, my bachelor thesis was about this research topic.
Summary
Cereblon protein (CRBN) is the receptor for substrates that bind to the CRL4CRBN E3 ubiquitin ligase complex, whose function is to control the selective degradation of proteins during various cellular processes. Some CRBN mutations associated with clinical cases of intellectual disability have been reported. One of these mutations occurred due to a change in Arg419, which generated a truncated version of the protein (R419X); and another mutation was caused by the substitution of the amino acid Cys391 by Arg in the CRBN binding site to ZN (C391R). At the moment, the genetic causes and phenotypic effects produced by CRBN mutations are known, but there is no information on the structural changes that cause these mutations.
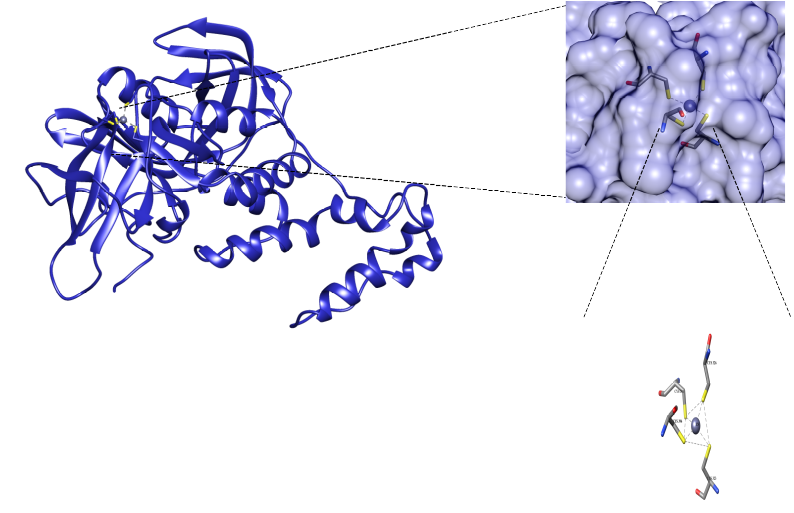
Figure 1.- CRBN protein and its ZN biding site. Retrieved from Ayala-Ruano, 2020, Bachelor’s thesis.
Therefore, in this research we used quantum mechanics (QM) and molecular dynamics (MD) simulations to study the changes caused by R419X and C391R mutations in CRBN. According to the results of the chemical parameters of the QM simulations, differences were observed between the native and C391R complexes in charge, angles, dihedral angles, and bond distances of the interacting atoms of the two systems, therefore the C391R mutation could affect the CRBN binding site to ZN. Considering the thermodynamic parameters of the QM simulations, it was observed that keeping the atoms of the Cys326, Cys323 and Cys394 residues fixed, the reaction would not be favorable, but leaving all the atoms free, the reaction could occur. Regarding the MD results, the simulations complied with the thermodynamic parameters, and there were differences in the conformational parameters (RMSD, Rg, and RMSF) between the native complex and the mutated systems. The results obtained showed that CRBN mutations could affect regions of the protein that bind to other substrates, and the fact that the substrates are not degraded could be one of the causes of intellectual disability in patients with C391R and R419X mutations.
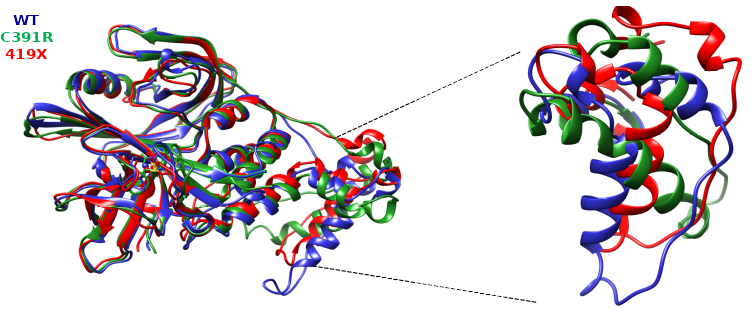
Figure 2.- 3D-superposition of CRBN protein and its mutant versions after molecular dynamics. Retrieved from Ayala-Ruano, 2020, Bachelor’s thesis.
Citation
Ayala-Ruano S (2020). Estudio computacional de los cambios estructurales de dos mutaciones de la proteína Cereblon. Bachelor’s thesis. Quito, Ecuador. url: https://repositorio.usfq.edu.ec/handle/23000/9950.
- Posted on:
- September 7, 2021
- Length:
- 2 minute read, 403 words